From Leaf to GTACCG: Sequencing DNA from Physocarpus
Botanists rely on external traits like leaf shape or flower size to make comparisons among plants and categorize them into taxonomic groups such as species or families. As a Ph.D. student in the Research and Conservation Department at Denver Botanic Gardens, I am using genetic information from DNA, in addition to morphology, to gain a deeper understanding about the evolutionary history of Physocarpus (“ninebark”). I’m a relative newcomer to genetics, and there’s still a certain magic to the idea that we can start with a leaf and “read” its DNA with sequencing technology.
The complete set of DNA in a cell is called the genome. Over long periods of time, changes in the pattern of DNA molecules (called nucleotides) accumulate within the genome. Geneticists can use these changes to make inferences about organisms by comparing DNA sequences from different specimens. In some cases, short sections of the genome are most relevant in addressing research questions (see the blog post What is DNA Barcoding?). In other cases, longer sequences are produced by stitching short segments together. This creates a closer representation of the whole genome. In my research with Physocarpus, I will be using a version of this approach called “double digest restriction site-associated DNA sequencing”, or ddRAD-seq for short.
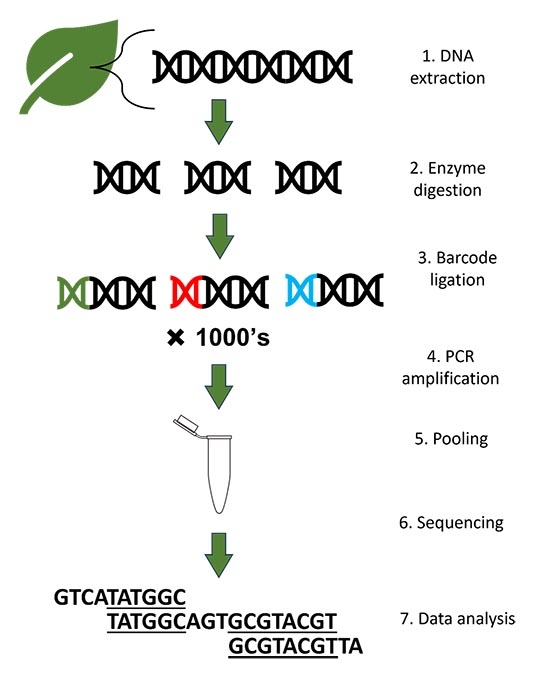
Generalized overview of DNA sequencing.
The first step of ddRAD sequencing is to get the DNA out of the plant in a process called DNA extraction. For vascular plants, DNA is most often extracted from dried leaf tissue. Second, the long strands of DNA are chopped up into small pieces with the help of specialized proteins called enzymes. This “enzyme digestion” process is necessary because DNA is a large and complex molecule and the technology capable of reading the whole genome doesn’t exist yet. Next, unique identifiers called “barcodes” are attached to the fragments. This is an important step because the fragments will be pooled together in one container before sequencing. The barcodes will help link each sequence back to the leaf sample from which it was extracted.
Midway through the DNA extraction protocol. Photo: Audrey Spencer
The next process, called amplification, makes lots of copies of the fragments in order to amplify the signal of the DNA. To help conceptualize this process, imagine one person shouting a message across a field; if hundreds of people were shouting the same thing, you’d be more likely to receive the message. In this analogy, the message is the signal from each nucleotide in the DNA sequence. Without amplification, the signal would be too low for detection.

Library preparation in the Genetics Lab. Photo: Andy Wilson
Next, the DNA fragments are pooled to create a “library.” A library contains millions of fragments of DNA from many leaf samples. It might not look like much (a small vial containing clear liquid), but the library is the product of months of work! The library is sent to a facility that has the specialized equipment required to “read” the sequences. Finally, the “reads” are returned electronically to the researcher. After some sorting and cleaning, the data are ready to be used in genetic analyses.
This post was contributed by Audrey Spencer, Ph.D. student in the Research and Conservation Department.
Add new comment